TEVC tips: working with mRNA
I wanted to share a few hard-earned tips for optimizing mRNA synthesis and mRNA injection for ion channel/receptor studies in Two-electrode Voltage Clamp electrophysiology (TEVC).
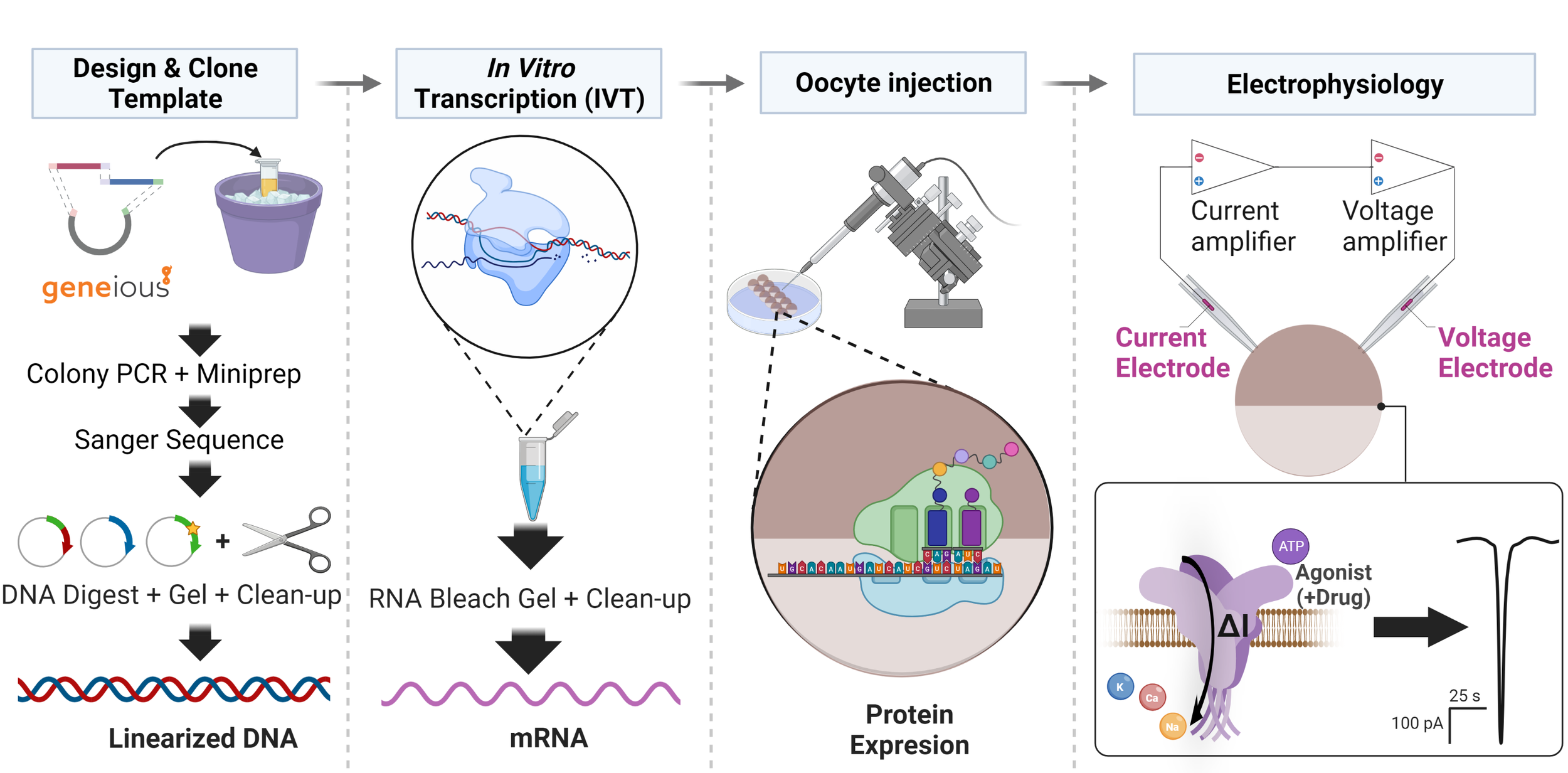
A workflow for a typical two electrode voltage clamp electrophysiology experiments. If all your oocytes keep dying, you won’t get data. If your oocytes live but can’t translate mRNA into protein (i.e. your RNA is degraded/contaminated or you’re using too many antibiotics) you don’t won’t get data. Triage and troubleshoot is the name of the PhD game in my opinion.
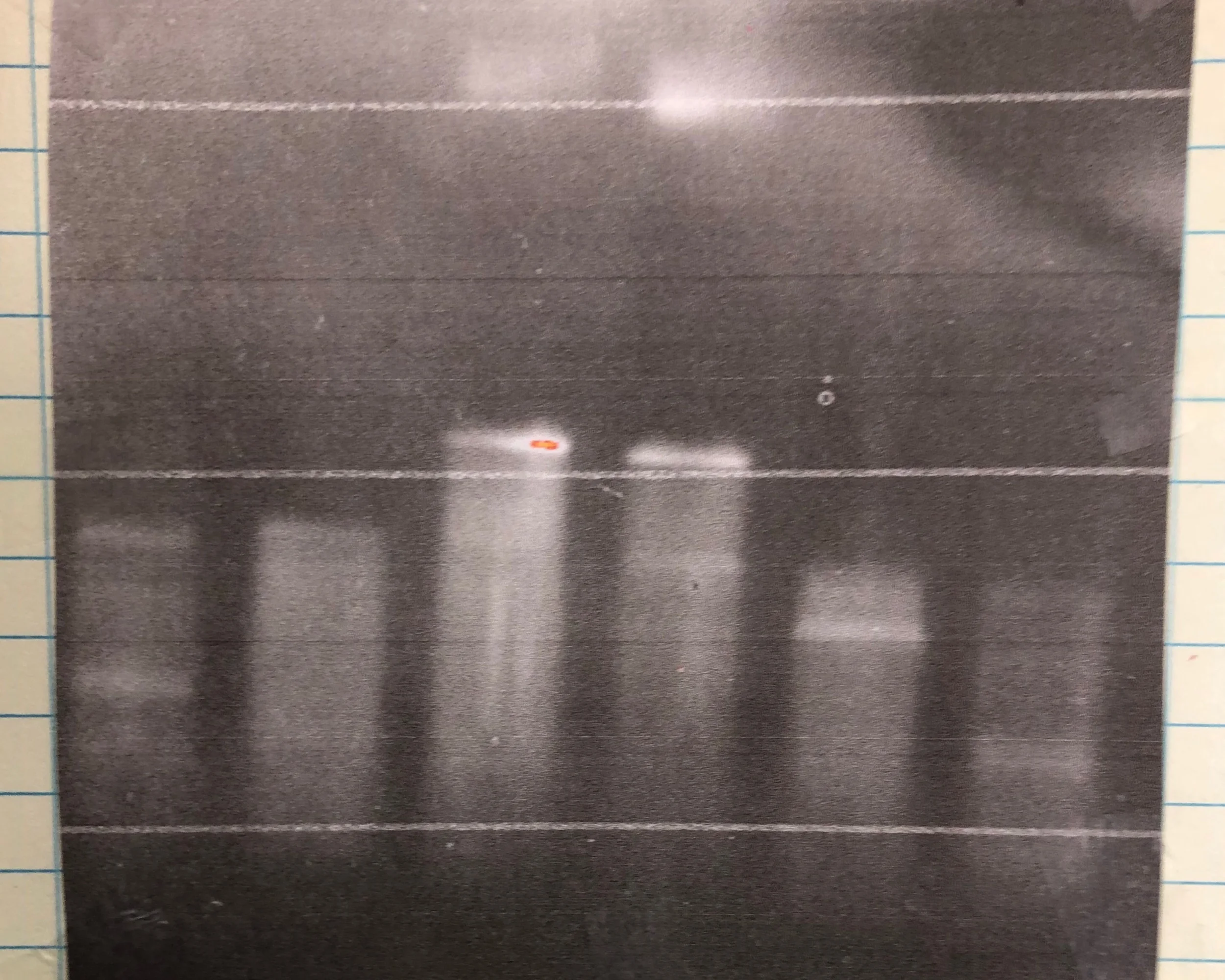
RNA bleach gel of my injection stocks back in February of this year. Lanes 4, 5, and 6 should all have 3 bands, but they’re not nearly as distinct as we want them to be.
(Some) peace of mind (from RNAses) in a container.
Gloves, always!!!!
Run restriction enzyme digests of your plasmid overnight, just so you’re sure that there’s no uncut carryover. Uncut plasmids could give you “off-target” RNA products.
For purifying linear DNA, I used to do precipitation with sodium acetate followed by ethanol extraction, but found that using Zymo’s DNA clean and Clear 5 kit works fine (no detectable RNAse activity.) Properly identifying a DNA pellet is tough if you’ve never done it before, and air-drying DNA (and RNA for that matter) always made me uneasy and seemed counter-intuitive to preventing contamination.
Always verify your digested DNA on a gel. 1 µL or less should be fine, since Ethidium bromide gels (with a gel imager) can detect > 50ng/µL of DNA. Using a UV light box is a little more difficult, but if you see a band run at the expected size, it obviously worked. If you don’t see a band, you either lost your product, or don’t have enough for RNA synthesis anyway. Consider diluting your RNA stocks with a buffer that contains RNAse inhibitor, like DEPC-treated water, or RNAsecure resuspension solution. I’ve used both, and have a small preference for DEPC water, just because the RNAsecure solution makes the buffer a bit “salty” which is sometimes evident by salt crystals forming when filling the injection needle
the RNAsecure solution can neutralize RNAses present in the solution if you heat it to 60°C for 15 min. I never used this feature but it may help out. Run an RNA bleach gel to verify your RNA synthesis results!!
If you’re performing the injections yourself (we use a nano-inject III), make sure you’re seeing consistent flow during the injection process.
The oocyte should visibly get bigger if injecting 20+ nL. The approximate volume of an oocyte is 1 µL, and injecting up to 50 nL (5%) should be safe.
Every 10-20 cells, inject RNA into the air, to check if the needle is clogged.
Needle puller settings do drift over time, so be aware that it may be time to readjust settings if your needle is too “wispy” or hair-like.
RNAse wipes. Use them to “sterilize” the environment you plan on working with during RNA preparation/injection.
Dedicated RNA-only tubes and pipette tips opened/used only for RNA preparation.
Store your injection stocks and stocks to dilute at -80°C.
Characterize your RNA synthesis yields and optimize your purification steps accordingly. For example, one of our receptors’ yield is never greater than 40 µgrams, which makes using Thermo’s Megaclear RNA clean up kit a little tough, since the minimum elution volume 50 µL. For this specific receptor, I use Zymo’s RNA clean up kit, which uses a 25 µL volume, and in my experience, is compatible with DEPC water. If the mRNA yield is less than 600 ng/µL I can’t use it for my studies, since I have two other receptors to co-express and dilute.
Note: Zymo does give out free samples if you want to try these out.
If you have to, you can always precipitate your RNA (with the LiCl in thermo’s T7/T3 mmessage mmcahine RNA synthesis kit) and alcohol-extract, for super-concentrated RNA yields. Trizol is also an option, although I’ve personally never done it.
An RNA bleach gel of my stocks back in May. Despite fairly distinct bands, nothing was expressing. The smearing we see isn’t outrageous, but remember that my plasmids/genes all lack a PolyA tail, and so they’re particularly susceptible to RNAse degradation.
An RNA bleach gel using 1 µL of RNA synthesized using the T7 mmessage mmcahine kit and purified using the thermo’s Megaclear kit. Everything was performed on the same day. You can see very distinct bands, although you also see very faint off-target products. These RNA products were synthesized using linear products containing a PolyA tail. The ladder on the left is the millennium RNA marker from Thermo, the ladder on the right is the NEB RNA ladder (old.)
Some Troubleshooting advice
One of the first RNA bleach gels I ever ran, circa 2016. You can see a lot of streaking and very little band definition. Some likely causes: contaminated tubes and tips, inexperience working with RNA, incomplete DNA digestion and/or clean-up. I definitely wore gloves though.
A relatively inexpensive oocyte injection dish. I never reuse oocyte injection dishes or the 3 mL pippettes we use to handle the oocytes, to reduce the likelihood of bacterial contamination. If you’re using a nano-inject III from drummond, you can really only do 50-60 oocyte i njections at a time before you have to refill the injection needle.
Inject an mRNA stock by itself, if the receptor is homomeric. If it’s expressing, then that receptor not the source of contamination. If it doesn’t express, it could be the source of the problem.
Don’t try to refill an injection needle if you’re injecting a lot of oocytes, or if you miscalculated the amount of mRNA needed to inject enough oocytes. By doing this, you run the rsk of contaminating the mRNA you plan to inject via the “used” injection needle. Bite the bullet and just set-up an entirely new needle, even if it’s only 10-20 oocytes.
Consider gel verifying your injection stocks (RNA bleach gel to the rescue.) This is especially true if you have an injection round or two that stops working; don’t continue to inject mRNA that doesn’t express.
Don’t make too large of an injection stock, in case it gets contaminated, and so that you don’t have to worry about how many freeze-thaw cycles the mRNA has undergone.
We stick to 15 µL, which lasts us 3-5 injection rounds of 30-50 oocytes (40 nL per oocyte.)
To prevent oocyte cross-contamination, consider make a new injection dish (each time if things are often bad). Fuse two small strips of a plastic mesh/sieve (1mm3 or smaller) to a 60 mm by 15 mm petri dish, either using super glue, a hot glue gun, or my preferred choice: 10-20 µLs of chloroform (melts the plastics together.) Be sure to rinse the injection dish well.
Before you pour an RNA gel, wipe down the combs with an RNAse wipe.
In my next post, I’ll cover a new method that fixed all my mRNA expression problems and accelerated my oocvte TEVC studies.