SLIC worked!
A quick update on my recent cloning experiment: it worked!
Fig. 1) A DNA gel I ran a few weeks back on the small SLIC fragment. The first lane is a DNA ladder (to approximate size), the next 3 lanes are the backbone (didn’t work, should see a large 8000bp product) and last 4 lanes are my insert (small, 750bp.)
The way to check whether your cloning experiment worked is usually by analyzing the DNA sequence of the product. You can always try to analyze the size of the product (faster but not definitive) on a DNA gel (See figure 1), and if you see a size similar to what you would expect, it’s probably what you’re interested in. However, at the end of the day, if you don’t analyze the DNA sequence, you can’t really be sure of what you have.
Sanger Sequencing
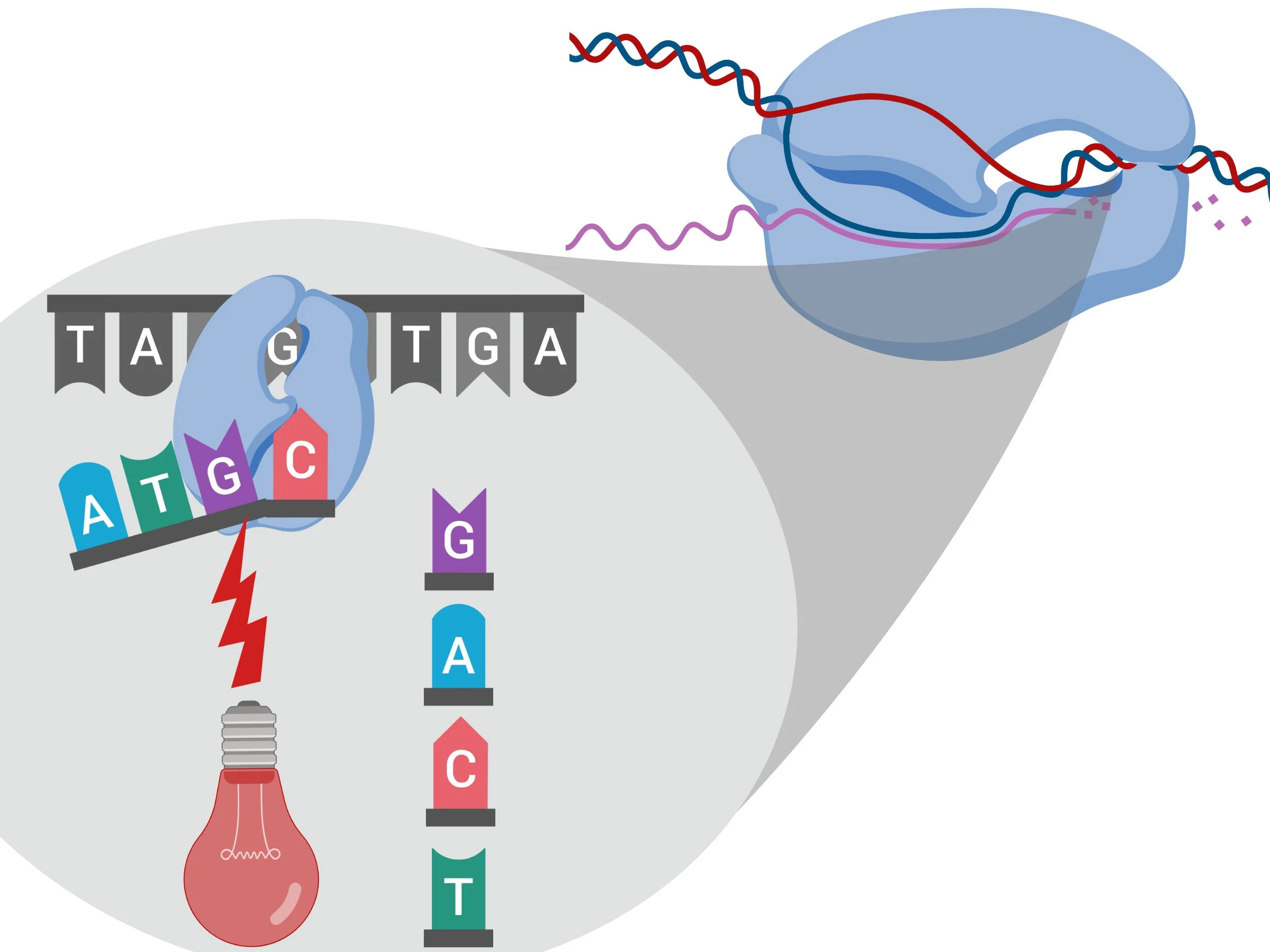
The way you sequence DNA is pretty cool: you take your regular (unlabeled) DNA product and add specifically colored DNA building blocks (i.e. nucleotides). For example, A (adenosine) might be colored blue, Cytosine might be colored red, etc.) You then add a primer, which is a short strand of DNA that is complementary to your DNA product ), and an enzyme called DNA polymerase (see figure 2).
Figure 2
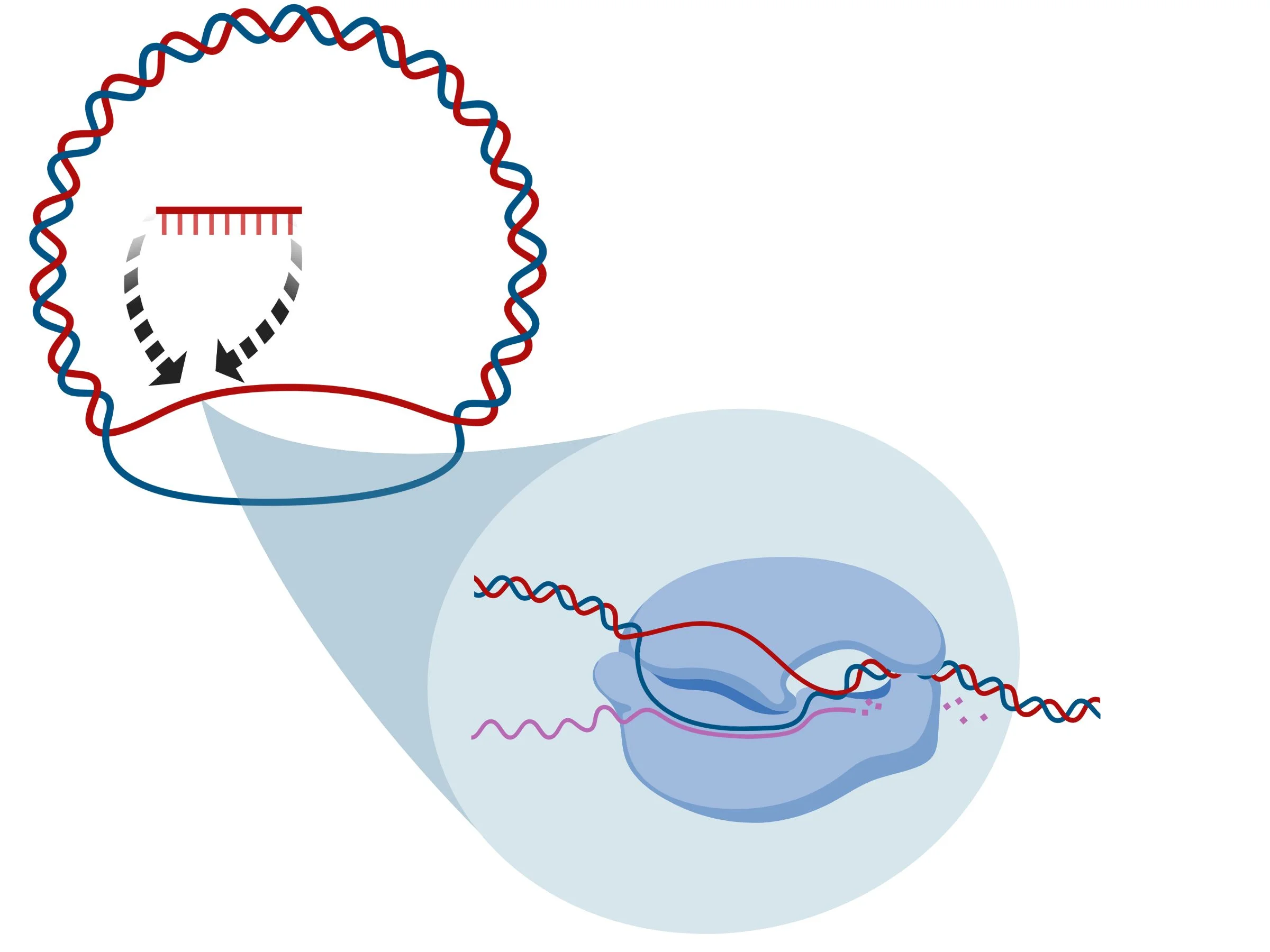
Figure 3
Figure 4
The enzyme amplifies the region targeted by your primer (see figure 3). While the DNA is getting amplified, you’re recording the colors (and therefore the sequence) of your target (see figure 4)
Results
Sequencing result with no problem
Example of a sequencing result that is “bad” or low quality (bottom reading), due to a weak signal.
Since it worked, now it’s time to scale up! I have about 25 combinations I want to try.
Conclusion
DNA sequencing used to be a very slow, expensive process, but in the past 40 years, advances in technology and molecular biology have made sequencing very cheap (a few pennies per base pair).
If you’re interested in the history of DNA sequencing, I highly suggest reading Dr. Leroy Hood’s biography, “Hood: Trailblazer of the Genomics Age” (see my thoughts on Instagram here). Like the title suggests, Lee Hood played a pivotal role in bringing in advances in DNA sequencing technology that allowed us to sequence the entire genome (the complete genetic material) of organisms, including humans.