Nanopores for Protein Sequencing
When I came across this new paper by Brinkerhoff et al, I knew I had to blog about it, and I finally had a chance to sit down and write about it. Cover art is the famous Oxford Nanopore MinION. Shoutout to the AAAS for granting me permission to use some figures from the paper! Per our agreement, “Readers may view, browse, and/or download material for temporary copying purposes only, provided these uses are for noncommercial personal purposes. Except as provided by law, this material may not be further reproduced, distributed, transmitted, modified, adapted, performed, displayed, published, or sold in whole or in part, without prior written permission from the publisher."
Multiple rereads of single proteins at single–amino acid resolution using nanopores
Conceptually, this paper amazing. Long-read/Single Molecule, Real-Time (SMRT) sequencing is an increasingly popular method in genomics, since one of the biggest advantages over the more traditional method (next generation sequencing) is that you don’t need to amplify or process your DNA sample for sequencing. You also have an incredible amount of creative freedom to set up experiments, the footprint in lab is small, and depending on your experimental setup, it might be cheaper than traditional NGS. Off the top of my head, a cool new thing to come out of this technology is plasmidsaurus, which is cheaper, faster, and more informative than the Sanger method for whole-plasmid sequencing.
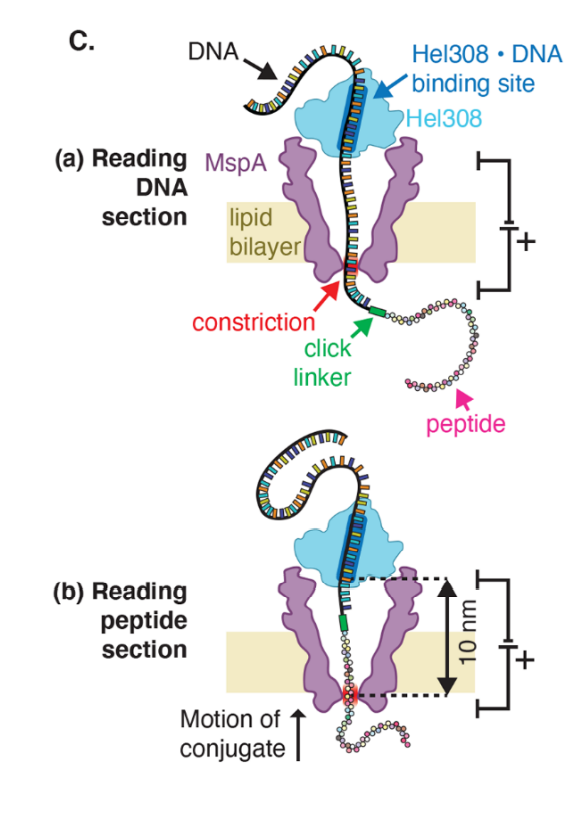
The way long-read sequencers (i.e. minION by Oxford Nanopore) work is that they step DNA through pores embedded in a (voltage-clamped) membrane. During the step process, the current will change according to the distinct electrostatic interactions caused by moving each nucleic acid through the membrane. Now, for this to work, you need to move DNA through the pore with the help of a motor enzyme, i.e. a DNA-protein interaction, and since protein-protein interactions are totally a thing, it makes sense to try to engineer a nanopore that works for proteins. In theory its possible, and in practice it can work, but not without some problems: early methods could only tell you the sequence of a single amino acid, not a peptide, and later methods could tell you the sequence of a peptide but due to the nature of the motor enzyme, results were difficult to interpret. With this in mind, Brinkerhoff et al set off to develop an improved system that could more accurately and precisely tell you the peptide sequence at the single amino acid level.
Constructs from H. Brinkerhoff et al., Science 10.1126/science.abl4381 (2021). Reprinted with permission from AAAS.
Schematic showing the sequencing process, from H. Brinkerhoff et al., Science 10.1126/science.abl4381 (2021). Reprinted with permission from AAAS.
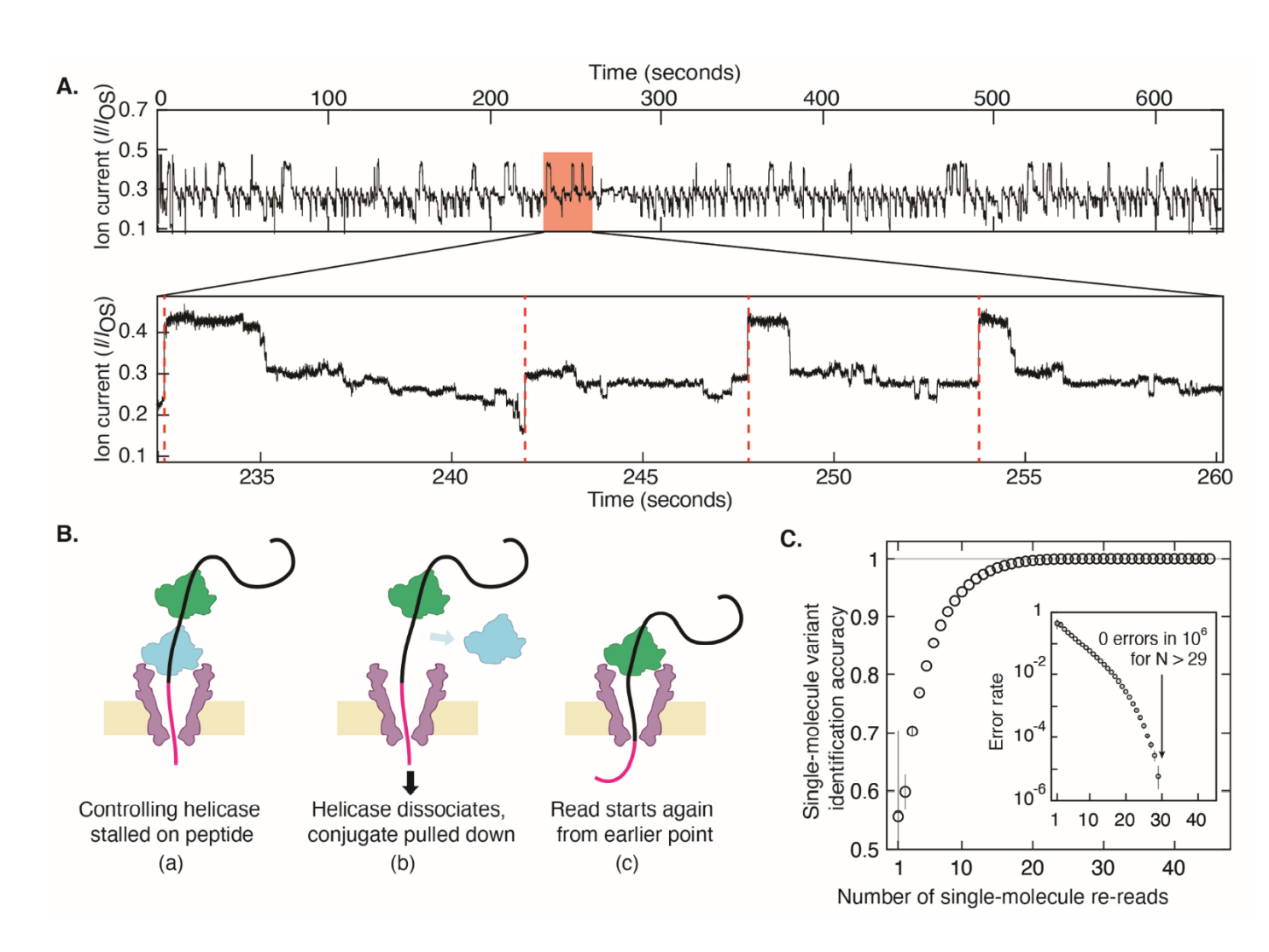
I’m not going to spend too much time summarizing the report, since it’s already well written and a quick read. What I will say is that it definitely a big step (pun!) for the field of proteomics. While it can’t (yet!) sequence proteins de novo (it can only sequence peptides linked to a DNA sequence) the fact that they’ve built a reliable method using commercially available reagents for accurately resolving the amino acids in a peptide is pretty freaking cool/exciting. Plus using their method, you can do multiple runs on your sample, which gives you less than 1 in 10^6 errors after 29 runs. Lastly, if you read a lot of ephys papers, you’ll recognize that some of the results look familiar (think single-channel current recordings and cumulative probabilities) because the data were generated on an Axopatch 200B amplifier and controlled with a National Instruments DAQ (see supplement for more info.) Absolutely nuts!
Sequencing results from H. Brinkerhoff et al., Science 10.1126/science.abl4381 (2021). Reprinted with permission from AAAS.
Examples of events, from H. Brinkerhoff et al., Science 10.1126/science.abl4381 (2021). Reprinted with permission from AAAS.
The proposal I submitted to Burroughs Wellcome Fund postdoctoral enhancement program in the first year of my postdoc originally combined electrophysiology with single-cell RNA [nanopore] sequencing and mass spec because 1) sequencing a neuron immediately after electrophysiology is a valuable technique (patch-seq!) 2) its lowkey inexpensive to play around with and 3) patching a neuron, collecting it in a recording electrode, and then (literally) turning around to prep and sequence the neuron yourself sounds hardcore/clever.
Lowkey, I’m hella pissed I didn’t get the grant because I would have 1000% added protein sequencing as a side project. I know I just said that this method not ready for primetime yet (mass spec is obviously still the major proteomics method, and I’ve been working on developing some cool ephys-proteomics methods since December of 2020) but protein nanopore sequencing is an area that’s about to heat up. I don’t think that’s clout chasing, its going where the action is ¯\_(ツ)_/¯